Exploratory Data Analysis
Learning objectives
- Identify data types and matching appropriate visualization techniques
- Explain the utility of exploratory data analysis
- Conduct exploratory data analysis on a new dataset
Why explore your data?
Why EDA?
- Check whether everything is as expected in your data
- Are there as many rows as you expected in the data set?
- Is any data missing?
- Are there data entry errors?
- Are there outliers?
Why EDA?
- Check whether everything is as expected in your data
- Check whether the assumptions of your modeling choice are met
- Do the data types match the analysis method?
- If doing simple linear regression is the relationship linear?
- If doing multiple linear regression, is the functional form modeled correctly?
- Are any points having a strong influence on the model results?
Reading in Data
Reading in data
Some data is already loaded when you load certain packages in R, to access these, you just need to use the data() function like this:
Reading in data
Other times you’ll have data in a file, like a .csv or Excel file. You can use read_* functions that load when you load the tidyverse package to read these in. For example, to read a .csv file in, you could run:
Note, movie_data.csv would need to be saved in your RStudio project folder for this code to run. We will practice this in a few weeks.
Checking your data
glimpse at your data
Rows: 1,846
Columns: 3
$ dataset <chr> "dino", "dino", "dino", "dino", "dino", "d…
$ x <dbl> 55.3846, 51.5385, 46.1538, 42.8205, 40.769…
$ y <dbl> 97.1795, 96.0256, 94.4872, 91.4103, 88.333…How many rows are in this dataset? How many columns?
00:30
glimpse at your data
Rows: 344
Columns: 8
$ species <fct> Adelie, Adelie, Adelie, Adelie, …
$ island <fct> Torgersen, Torgersen, Torgersen,…
$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3…
$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6…
$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181…
$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650…
$ sex <fct> male, female, female, NA, female…
$ year <int> 2007, 2007, 2007, 2007, 2007, 20…What type of variable is species? How many numeric variables are there?
00:30
Visualizing data
What are we looking for?
- The “shape” of the data
- Patterns
- Outliers (strange points / data errors)
Data
Let’s grab one of the datasaurus_dozen datasets.
What does filter do? Why ==?
00:30
One continuous variable
Histogram
The geom_* in ggplot2 describe the type of plot you want to create. What do you think would create a histogram?
00:30
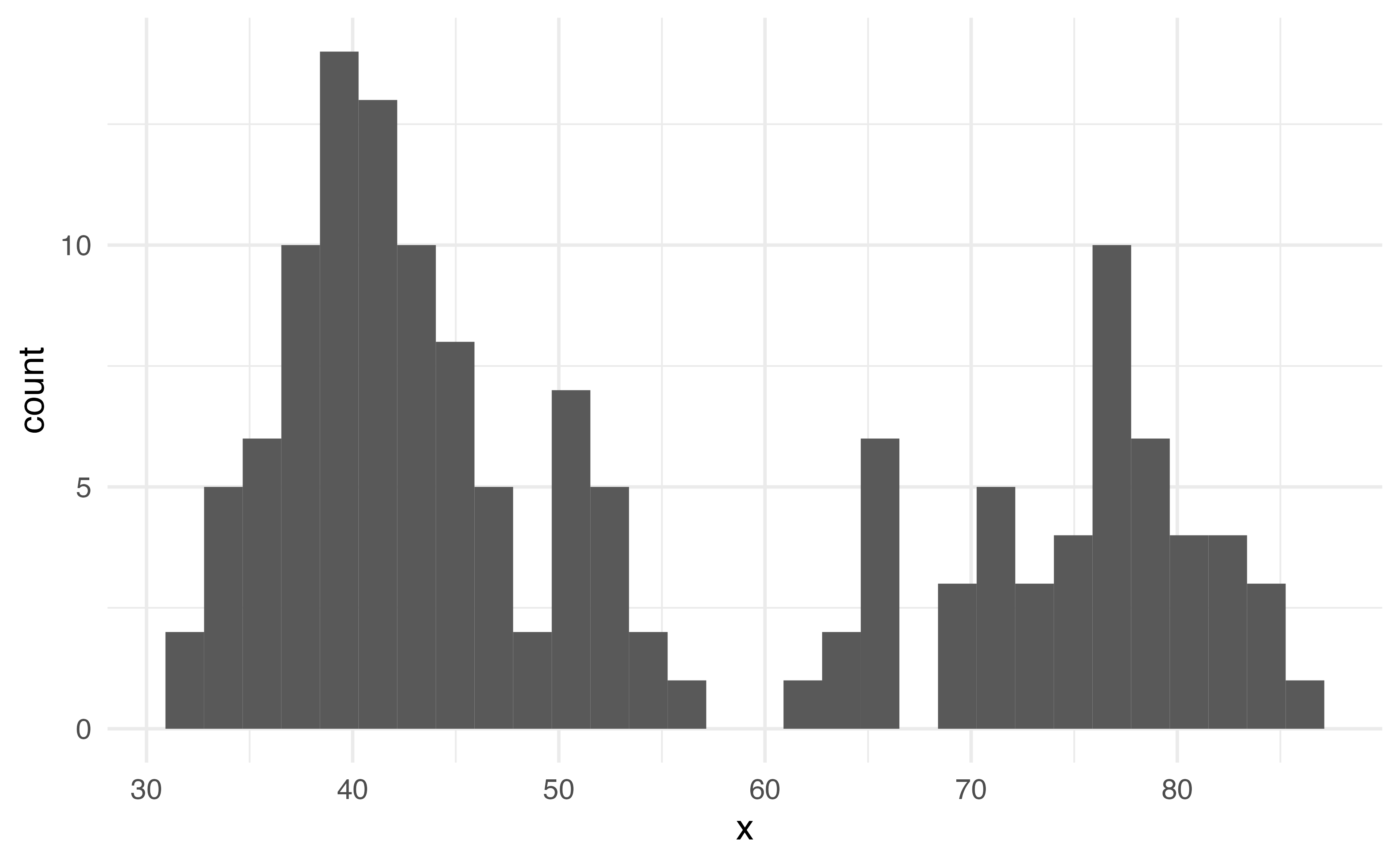
geom_histogram
Histogram
What does this warning mean? How do you think we can get rid of it?
Histogram
Histogram
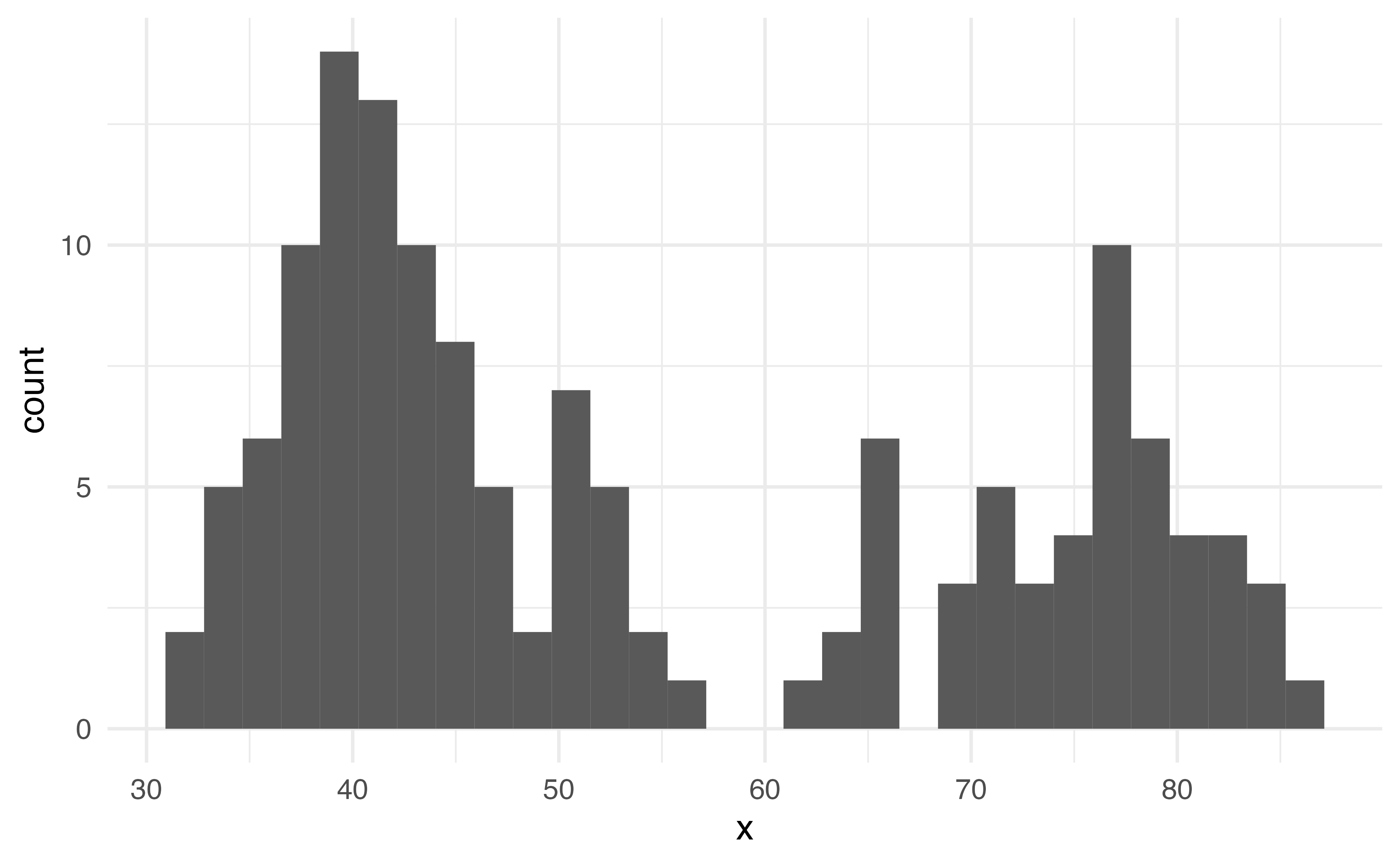
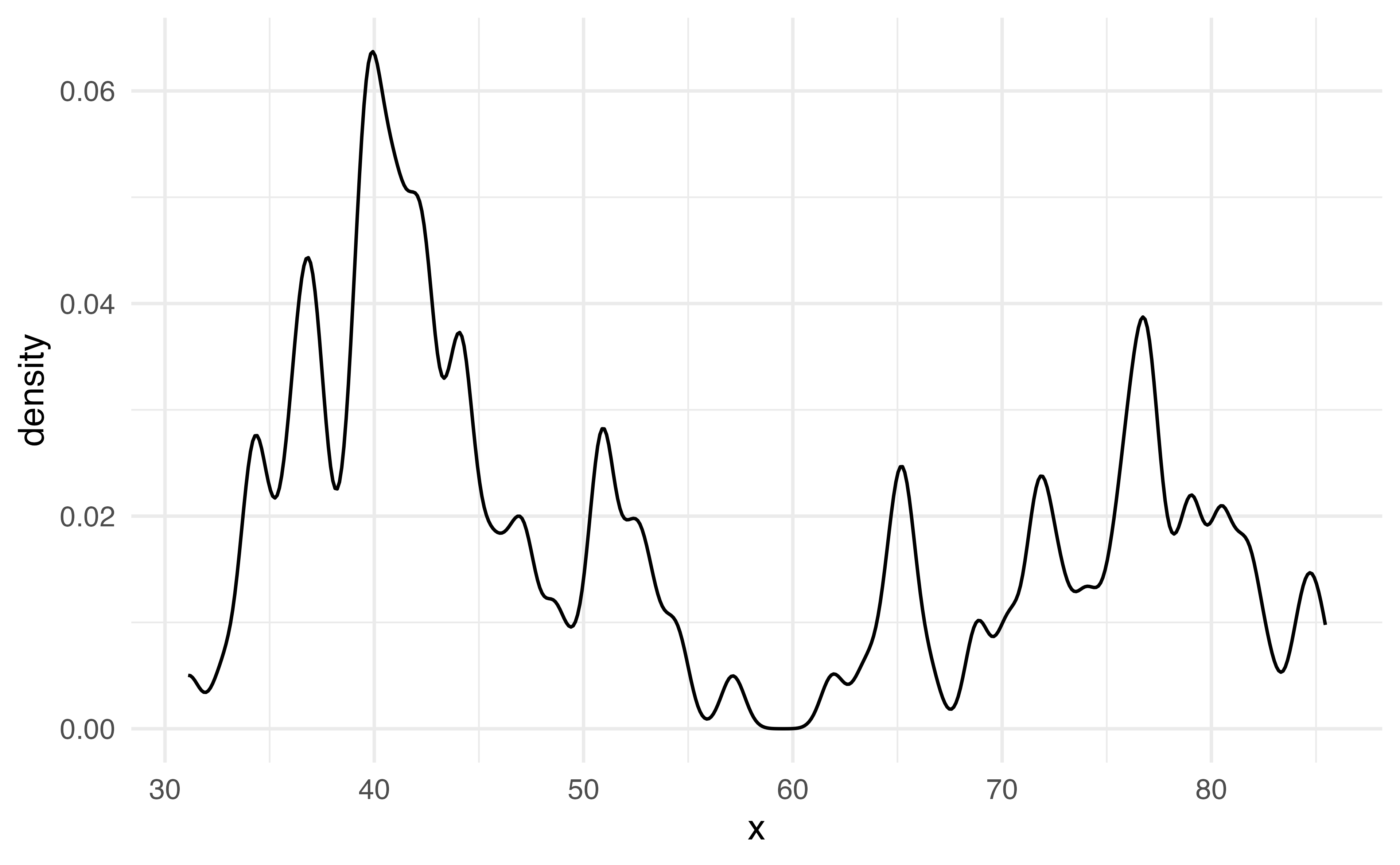
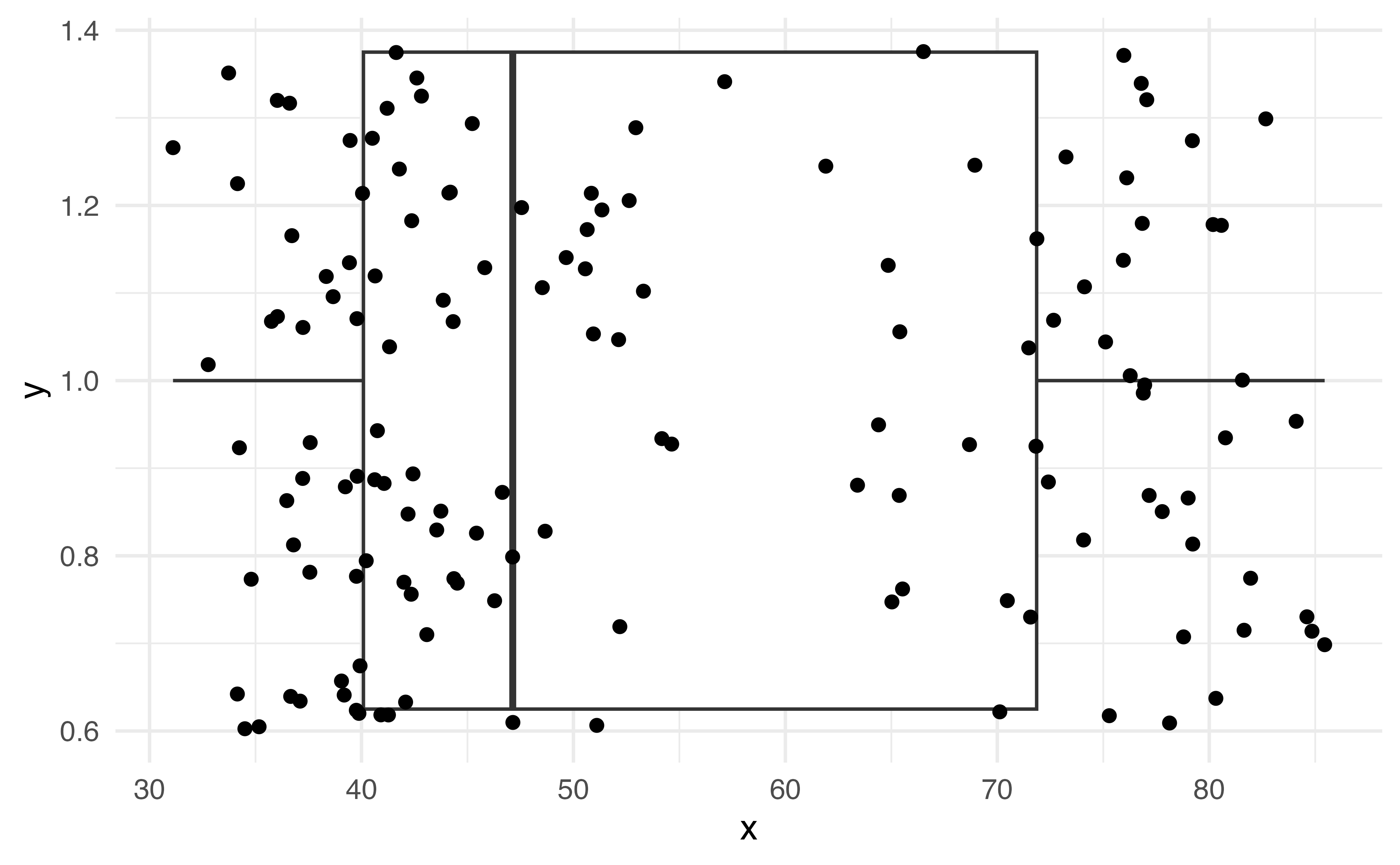
What does this plot tell us about the shape of this data?
00:30
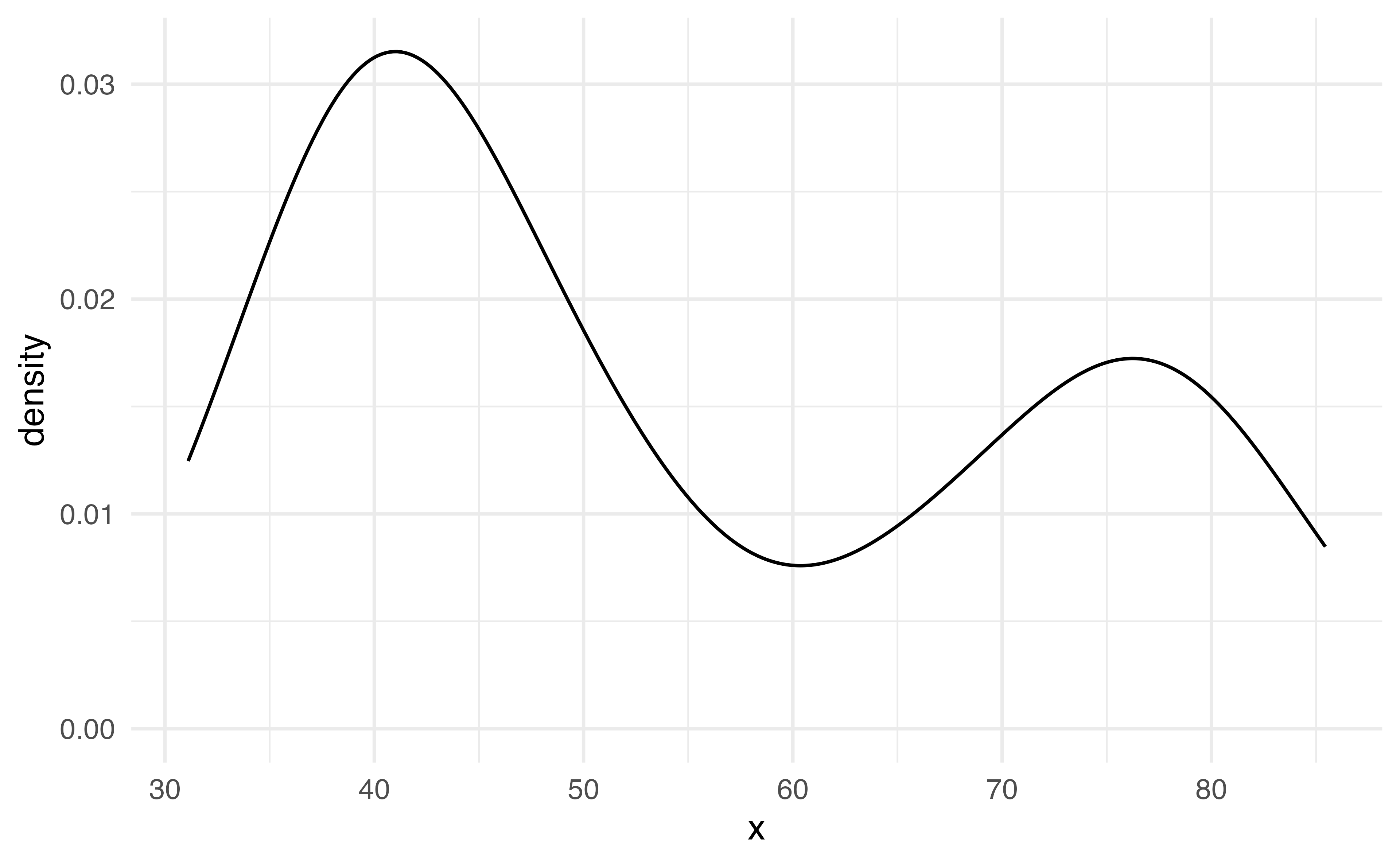
Density plot
What geom_ do you think would create a density plot?
00:30
geom_density
Density plots
Density plots
Boxplot
What geom_ do you think would create a boxplot?
00:30
geom_boxplot
Boxplot
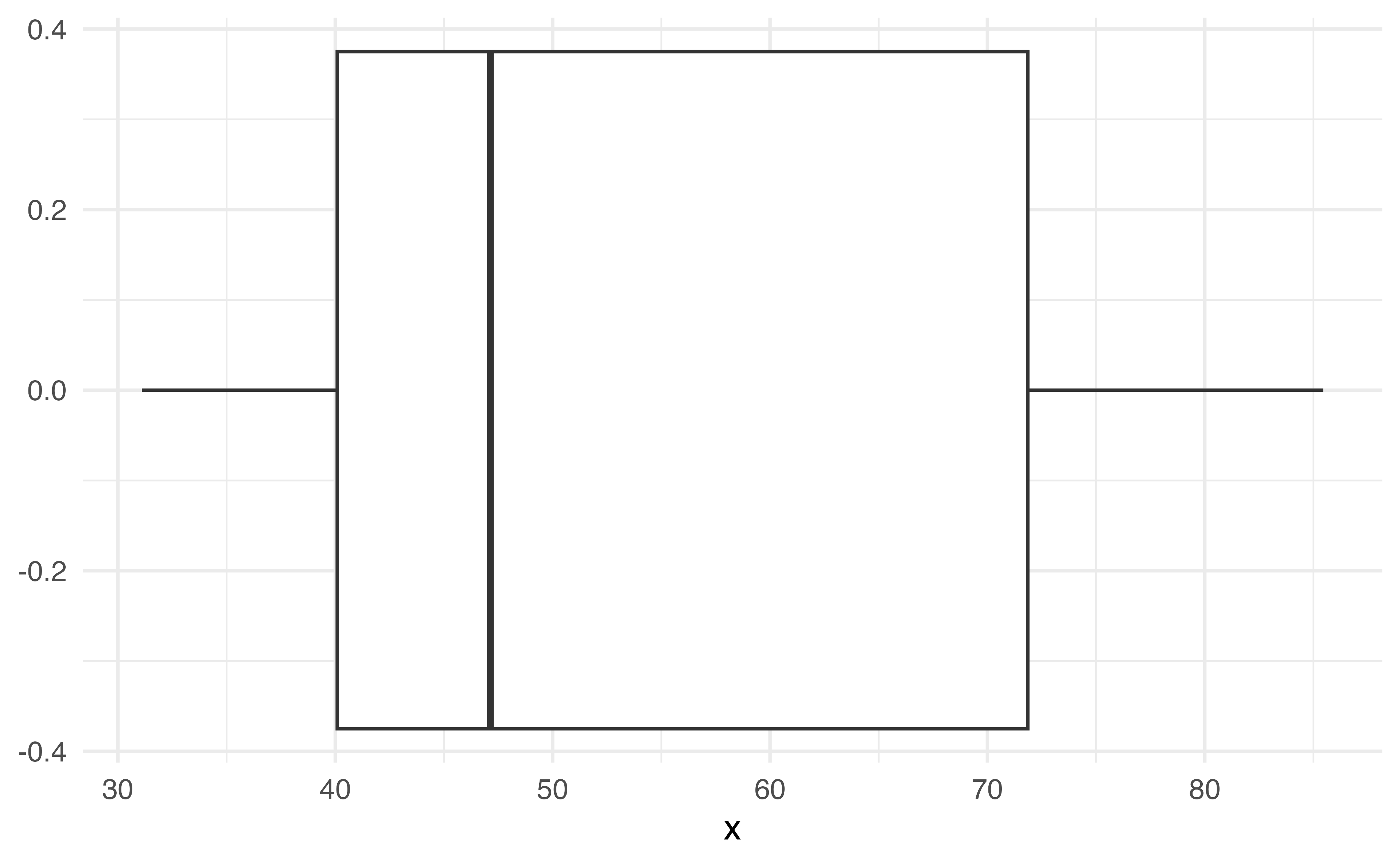
Does this give us as much information as the histogram?
00:30
Boxplot
Always show your data!
Boxplot
Always show your data!
Relationship between two continuous variables
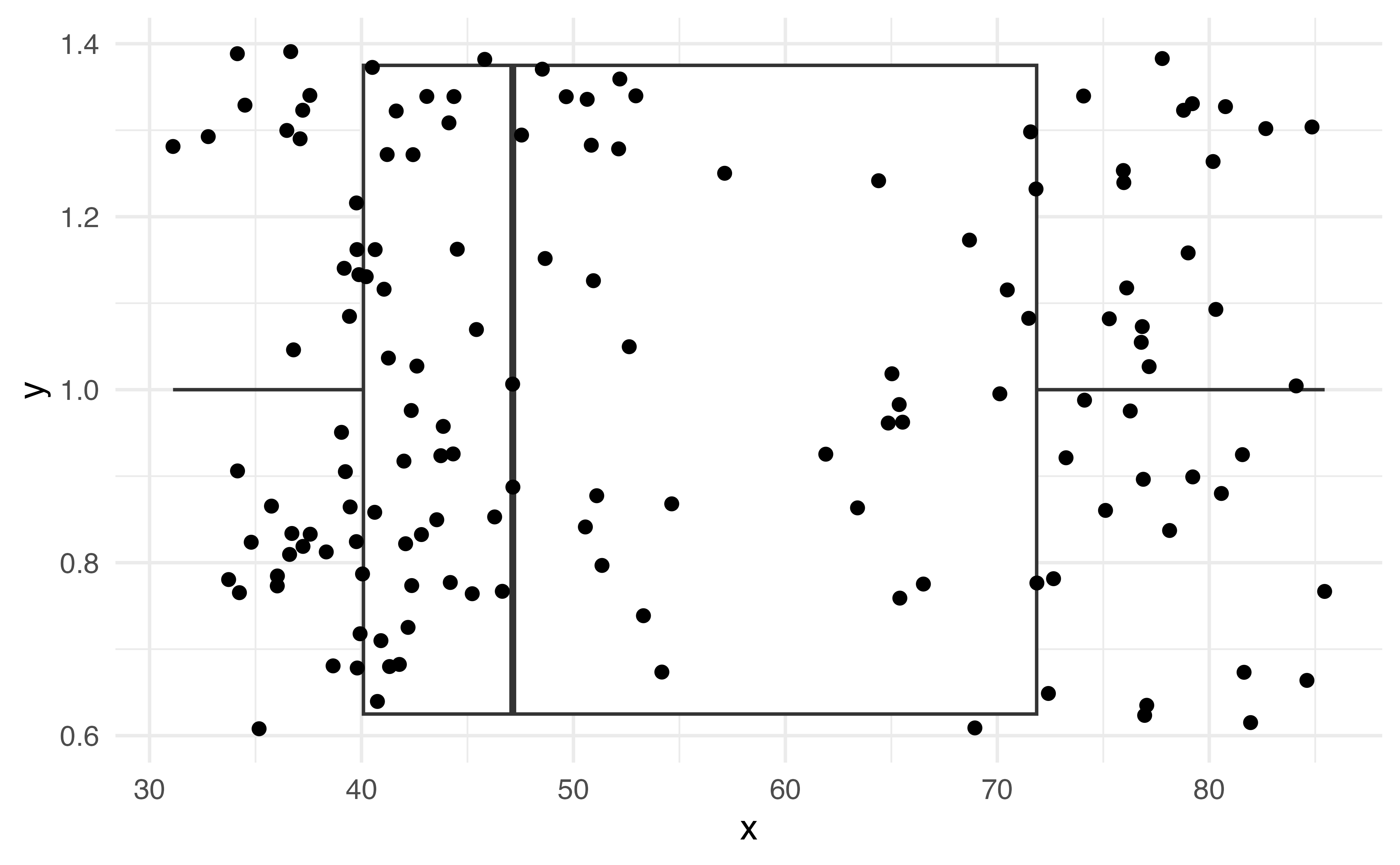
Scatterplot
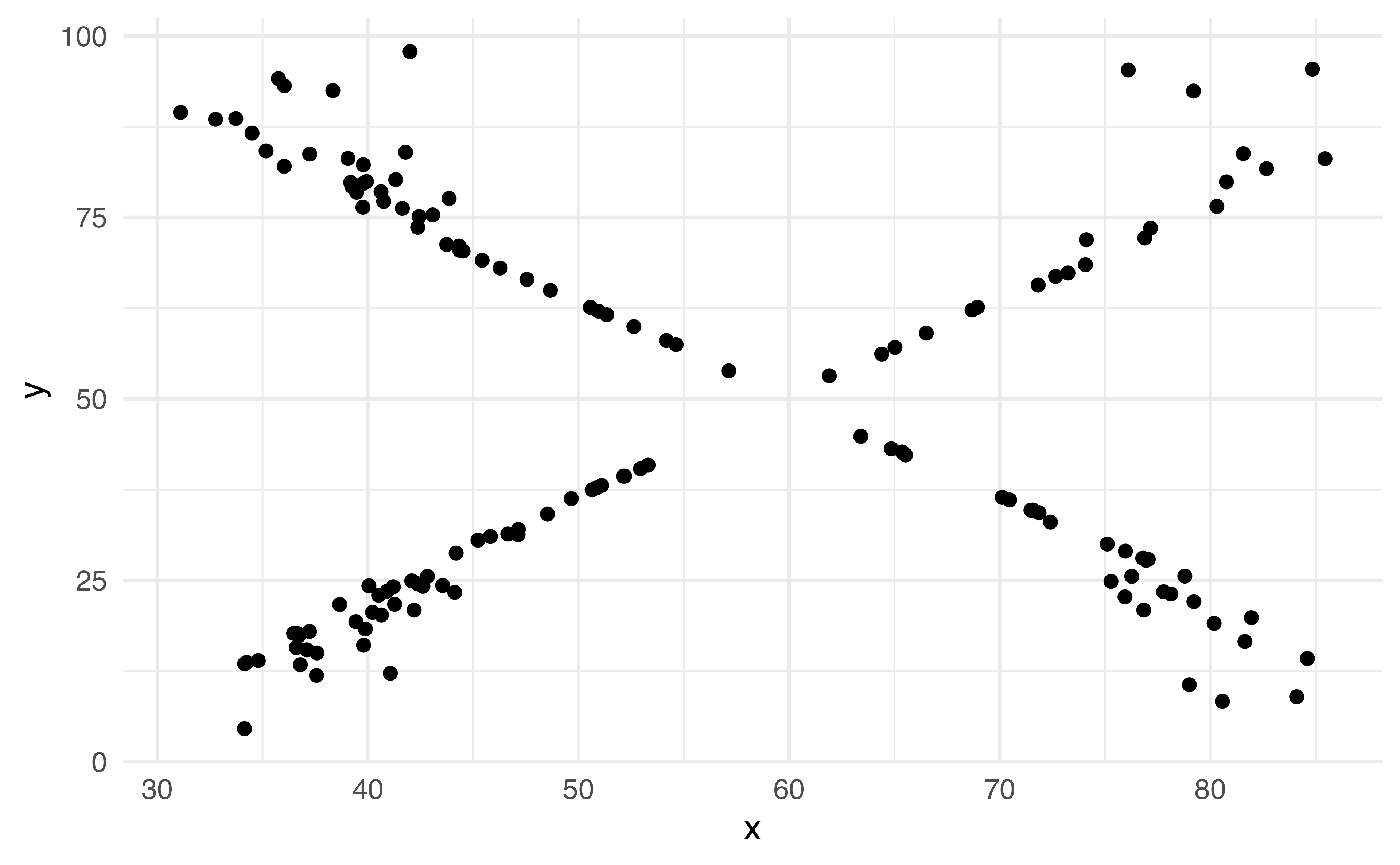
Hex plot
One categorical variable
Barplot
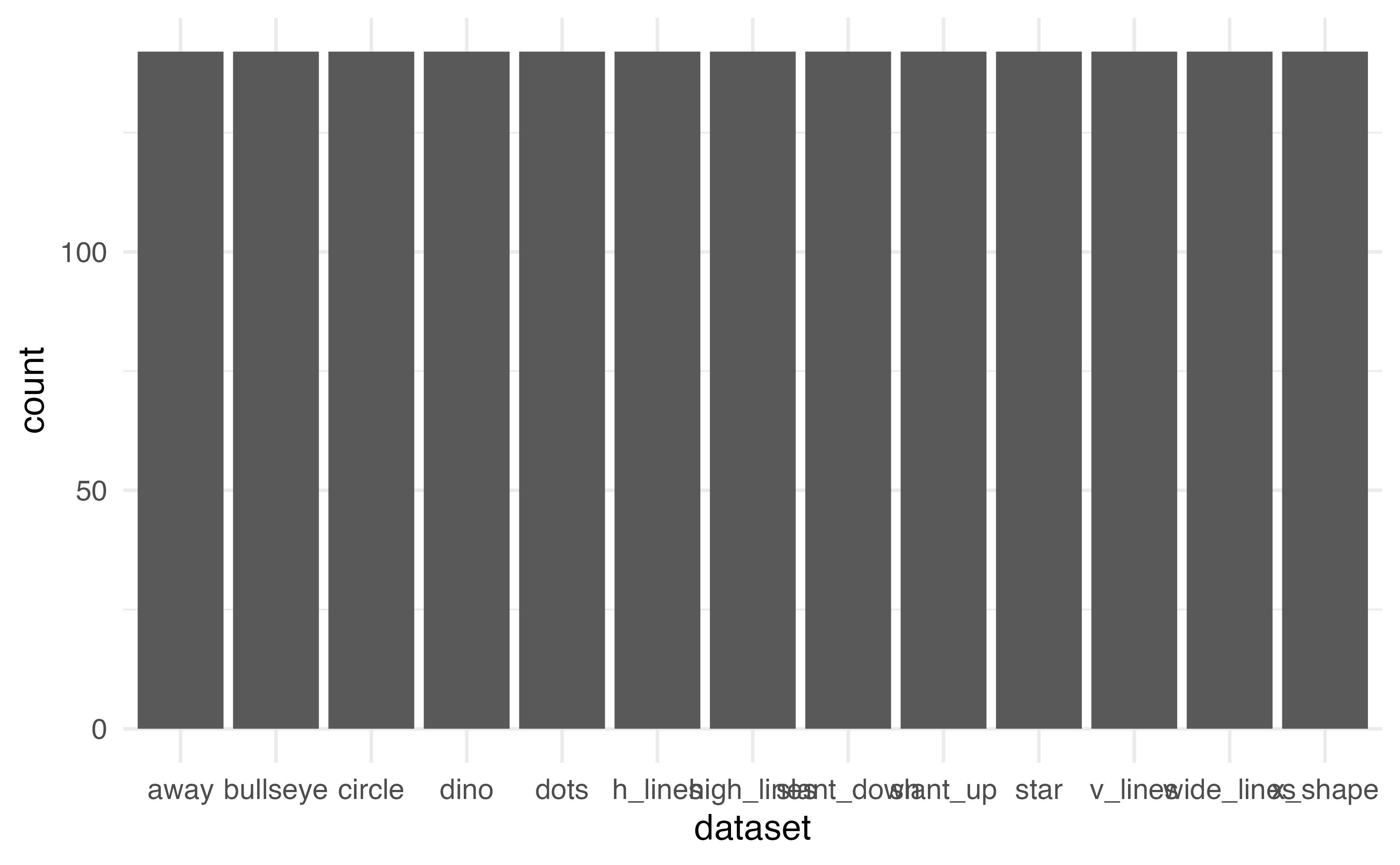
What does this plot tell us?
00:30
Barplot
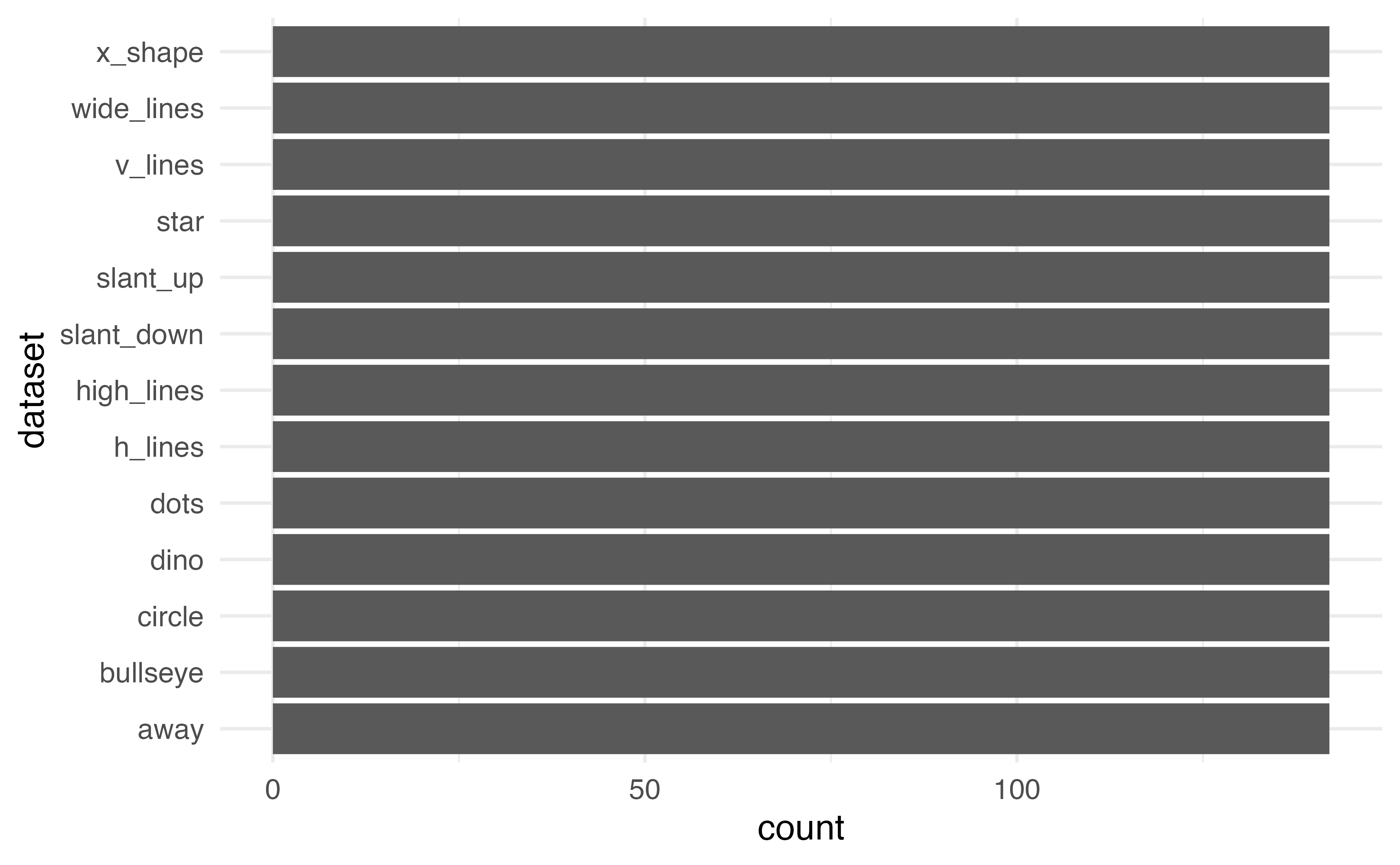
Flip the coordinates to make it easier to read
Relationship between continuous and categorical variables
Histogram
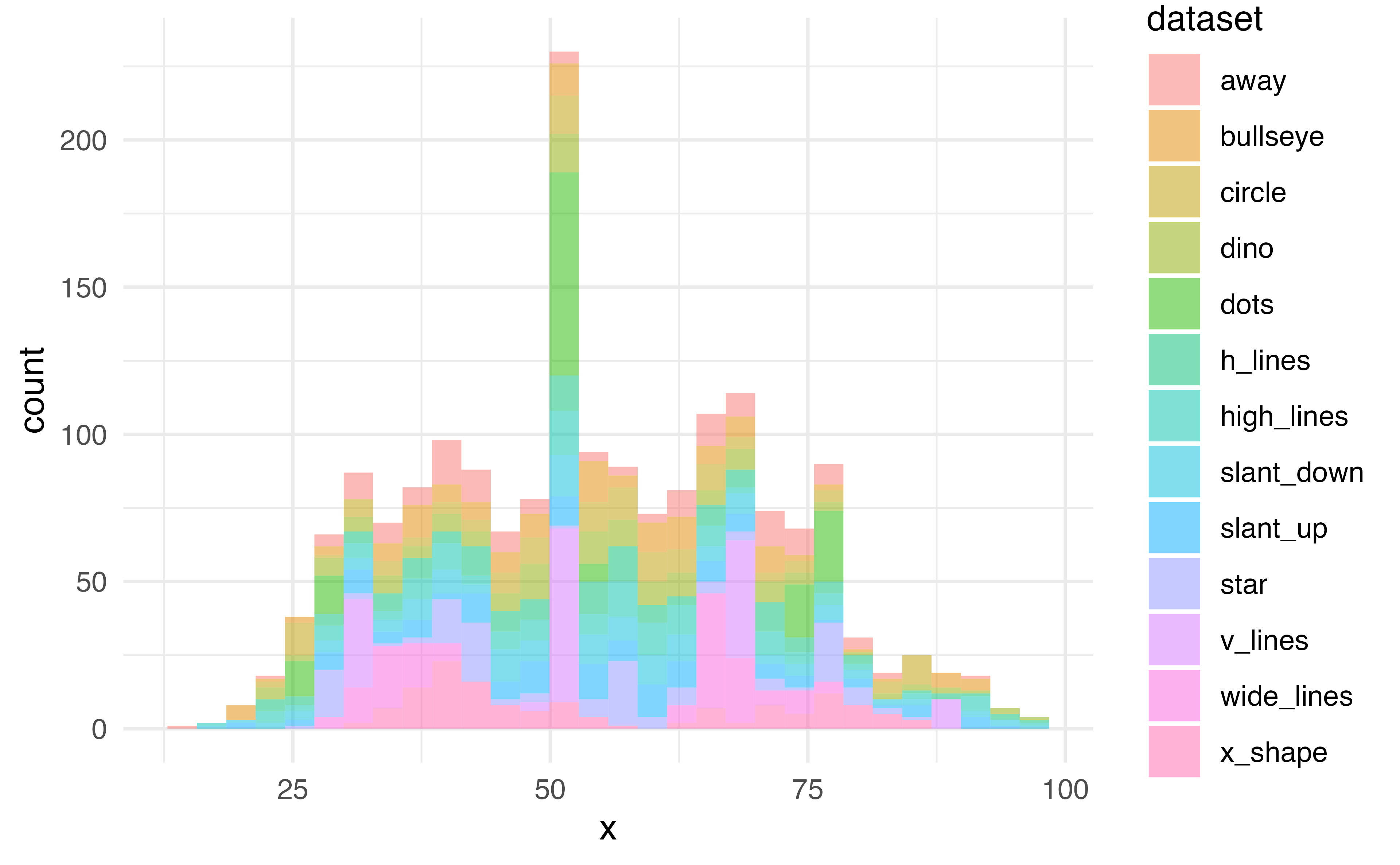
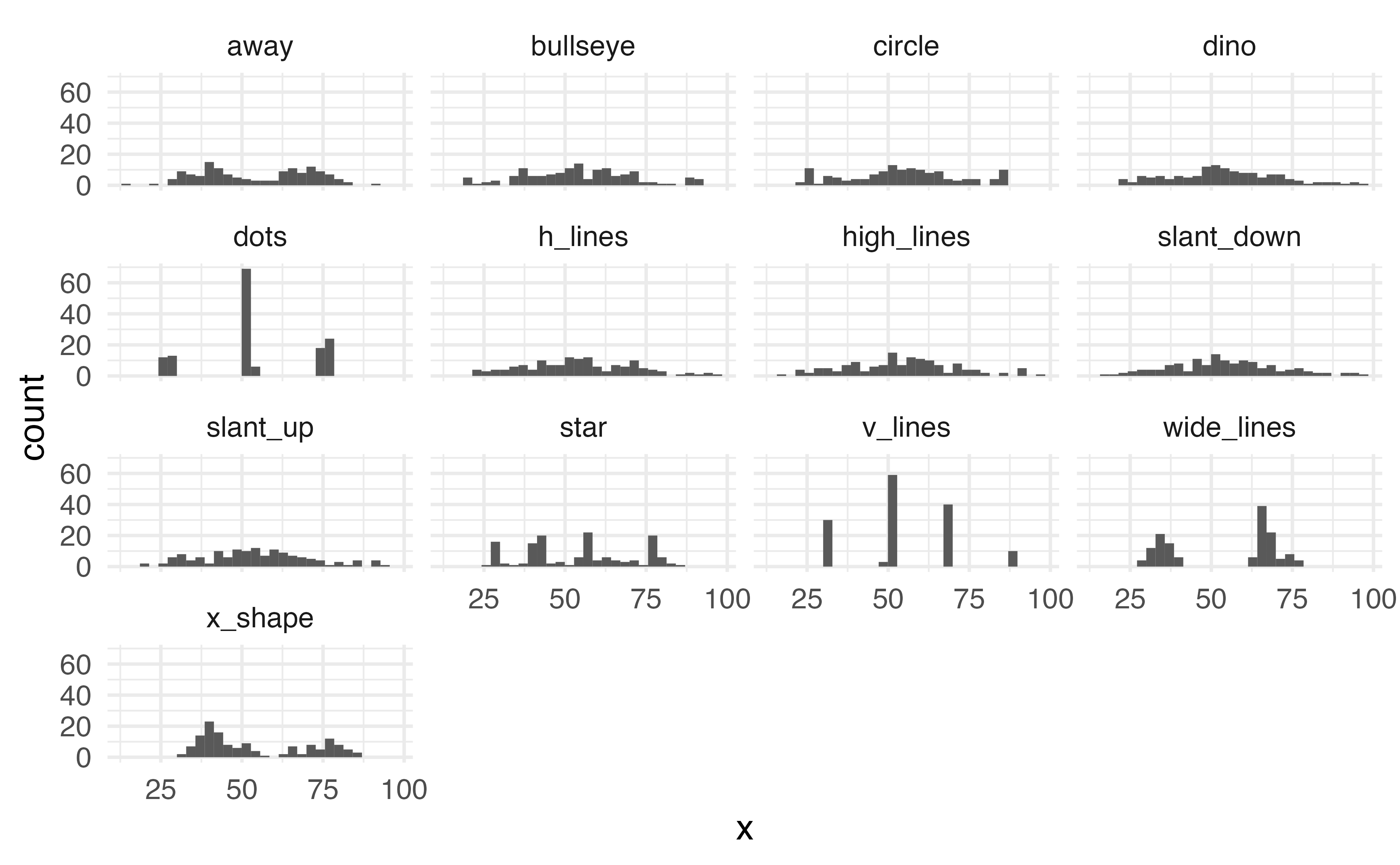
Histogram
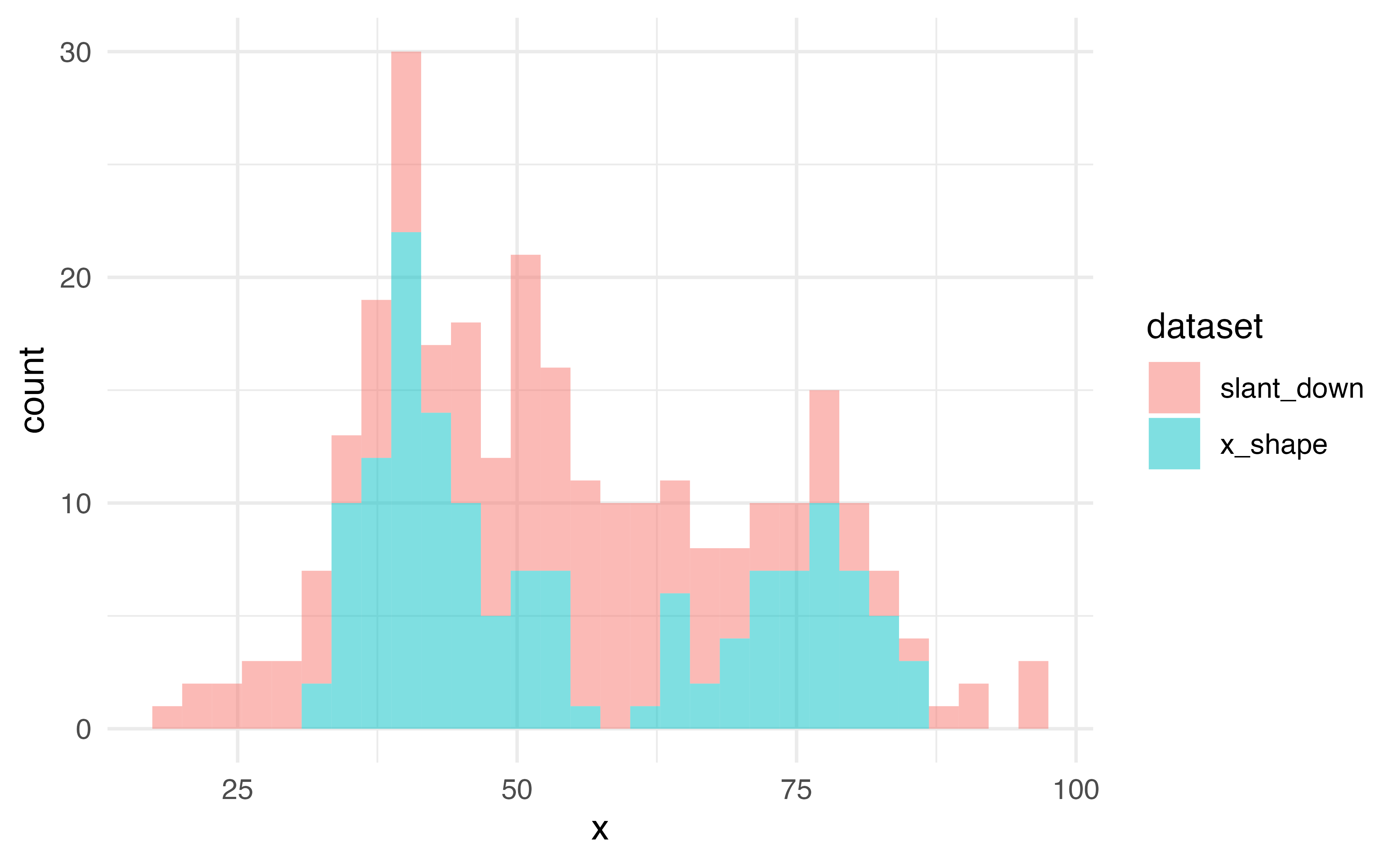
Histogram
What does %in% do?
00:30
Histogram
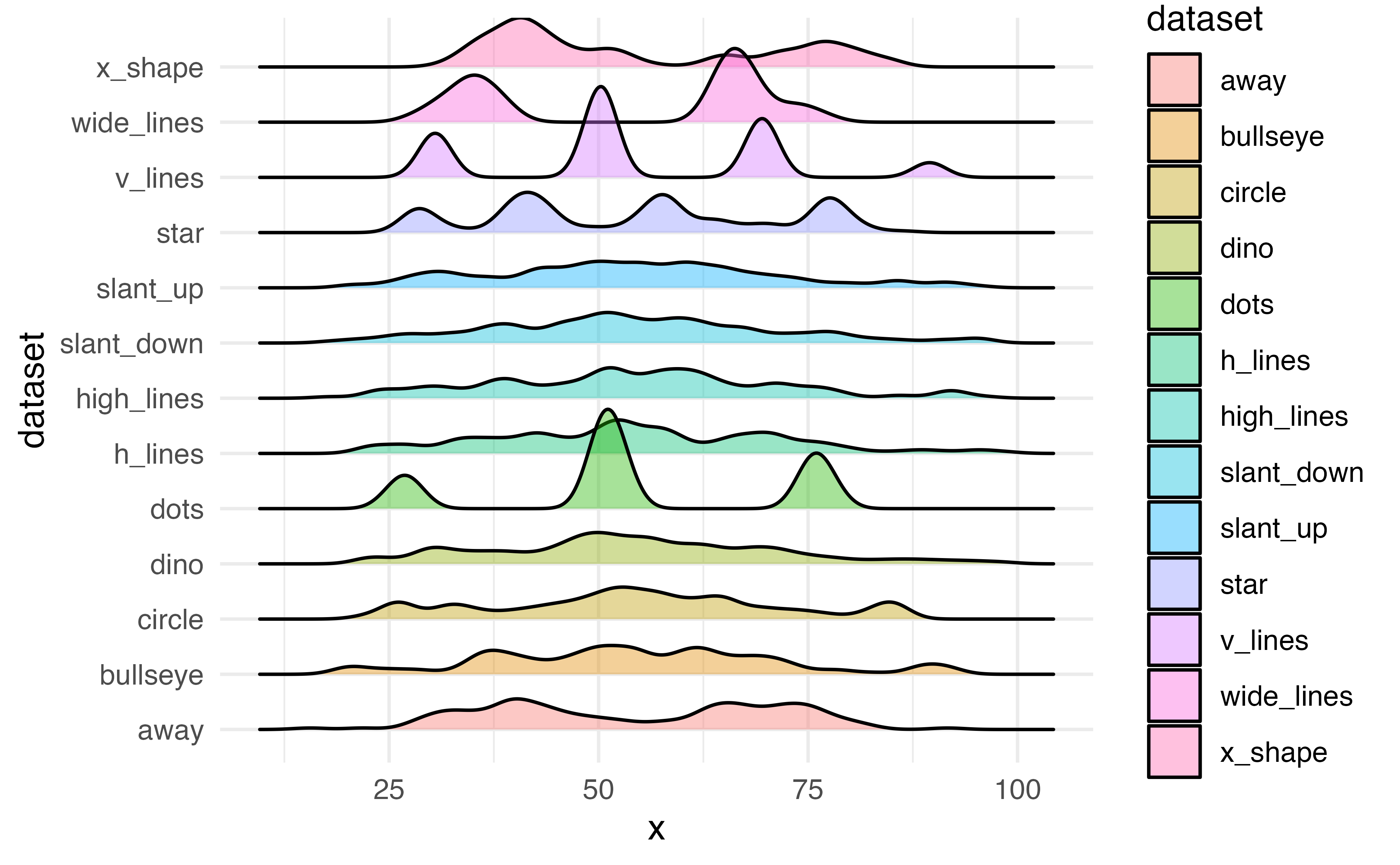
Ridge plots
Boxplot
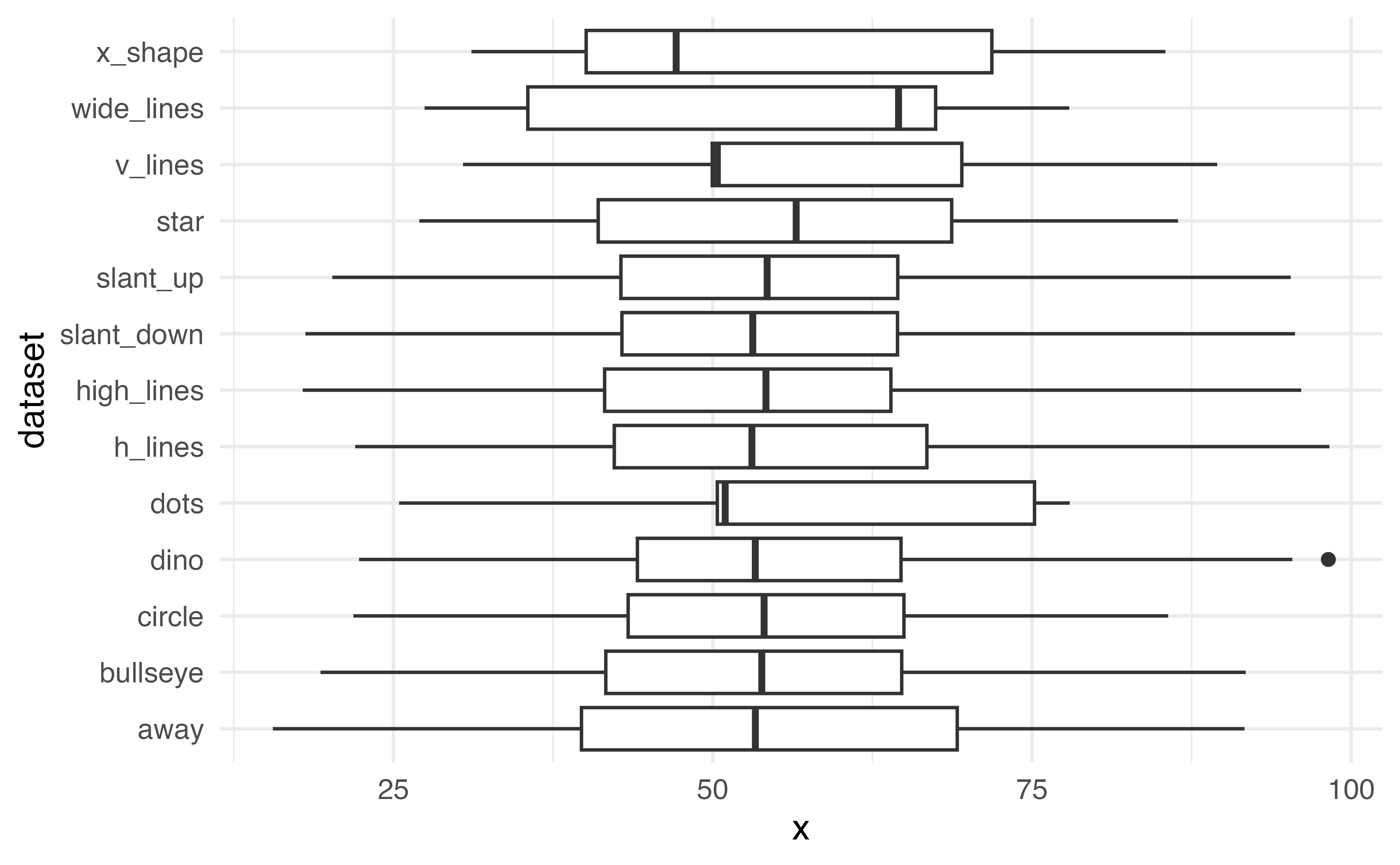
What is missing?
00:30
Boxplot
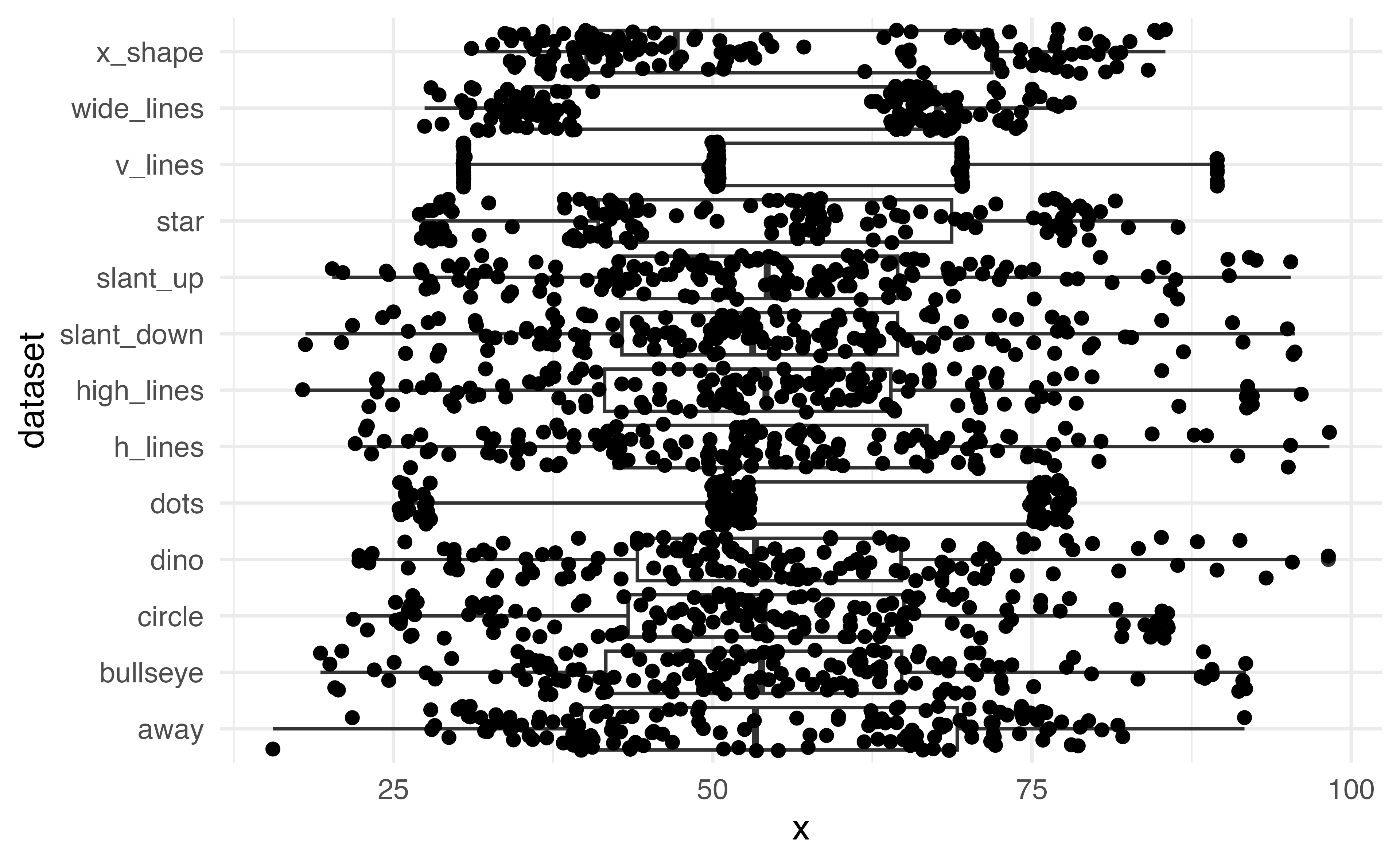
How can we make this more legible?
00:30
Boxplot
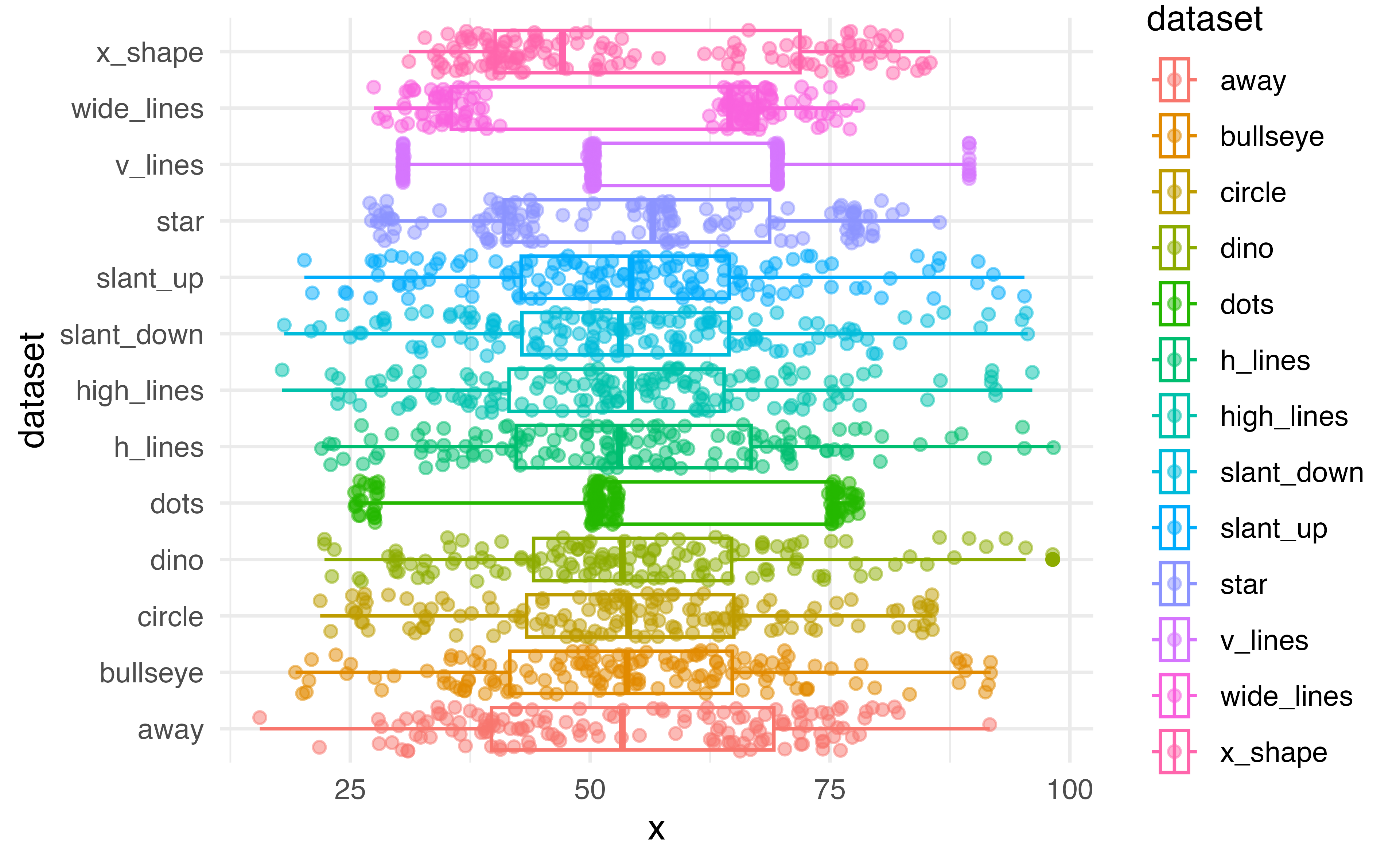
How will we use this?
- Plot every outcome variable before performing an analysis
- Be sure to include labels and titles on all plots for full points
- Plot important features
- Be sure to note any missing data patterns
Application Exercise
Open the Welcome Penguins folder from the previous application exercise
Create a boxplot examining the relationship between the body mass of a penguin and their species.
Add jittered points to this plot
Add labels and a title to this plot
08:00