01:00
Random Forests
Random forests
Do you ❤️ all of the tree puns?
Random forests provide an improvement over bagged trees by way of a small tweak that decorrelates the trees
By decorrelating the trees, this reduces the variance even more when we average the trees!
Random Forest process
Like bagging, build a number of decision trees on bootstrapped training samples
Each time the tree is split, instead of considering all predictors (like bagging), a random selection of \(m\) predictors is chosen as split candidates from the full set of \(p\) predictors
The split is allowed to use only one of those \(m\) predictors
A fresh selection of \(m\) predictors is taken at each split
typically we choose \(m \approx \sqrt{p}\)
Choosing m for Random Forest
Let’s say you have a dataset with 100 observations and 9 variables, if you were fitting a random forest, what would a good \(m\) be?
The heart disease example
Recall that we are predicting whether a patient has heart disease from 13 predictors
1. Randomly divide the data in half, 149 training observations, 148 testing
2. Create model specification
mtry here is m. If we are doing bagging what do you think we set this to?
2. Create bagging specification
What would we change mtry to if we are doing a random forest?
2. Create Random Forest specification
- The default for
rand_forestisfloor(sqrt(# predictors))(so 3 in this case)
3. Create the workflow
4. Fit the model
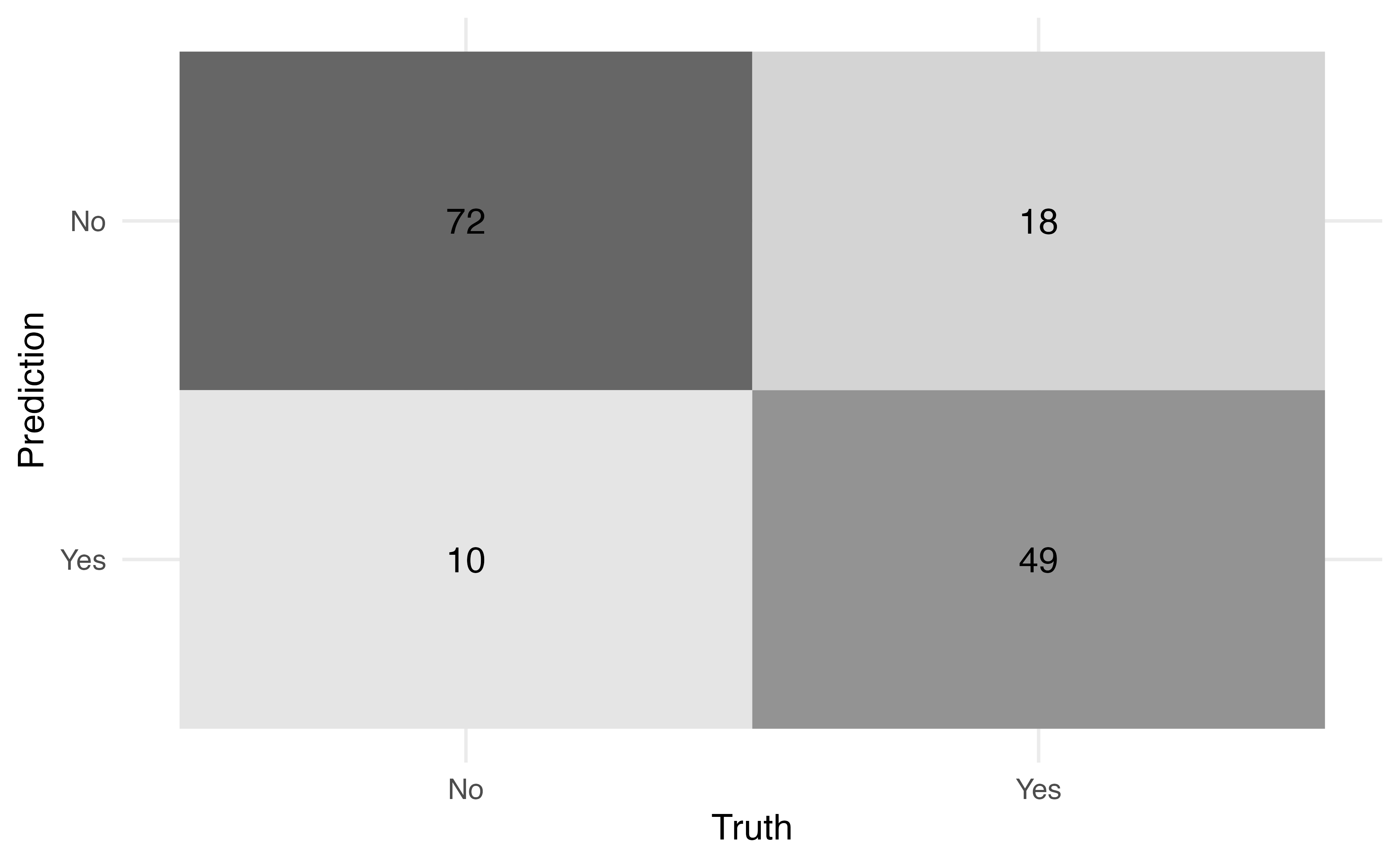
5. Examine how it looks in the test data
Application Exercise
- Open your last application exercise
- Refit your model as a random forest
10:00
Dr. Lucy D’Agostino McGowan adapted from slides by Hastie & Tibshirani